Promoter Analysis
(examination of the binding sites of Ahr1 using ChIP-seq data from Candida albicans)
(Version: April 2021)
Background
Candida albicans is a human pathogenic fungus which can cause severe infections in immunocompromised people. It can grow in different variations: as unicellular yeast, pseudohyphae and hyphae [1]. This morphological flexibility and especially the hyphal form are crucial for the virulence of the fungus [2, 3]. In recent years, several virulence factors of C. albicans have been identified. They are often closely connected with hyphal growth. This includes the transcription factor Ahr1 which is supposedly involved in the regulation of several virulence associated genes via binding their promoter region.
"Chromatin Immunoprecipitation sequencing" (ChIP-Seq)
Two C. albicans strains with a hyperactive Ahr1 were analyzed using ChIP-seq [4]. This procedure can determine protein-DNA-interactions. The protein is mixed with the DNA, the DNA is fragmented and all unbound DNA pieces are removed. Afterwards, the protein is washed off the DNA and the sequence fragments are analyzed using high throughput sequencing. The result of this analysis were 325 sequence fragments representing potential binding sites of Ahr1 in the C. albicans genome. By using the chromosomal position of those fragments, 532 genes on both DNA strands were identified that harbored the sequences in their promoter regions meaning they could be regulated by Ahr1.
Promoter Analysis
A region of 500 base pairs around the maximum of each peak of a sequence fragment (which is the most abundant location of this fragment in the high throughput sequencing and therefore the most probable binding site) was used as input for the online tool Meme-ChIP (v. 5.1.0) [5]. This way, a highly significant motif was found that is concurrent to an already known binding motif of Ahr1. For further confirmation, the software MochiView (v. 1.46) [6] was used. The integrated motif finder also detected the known Ahr1 motif, which is displayed in figure 1.
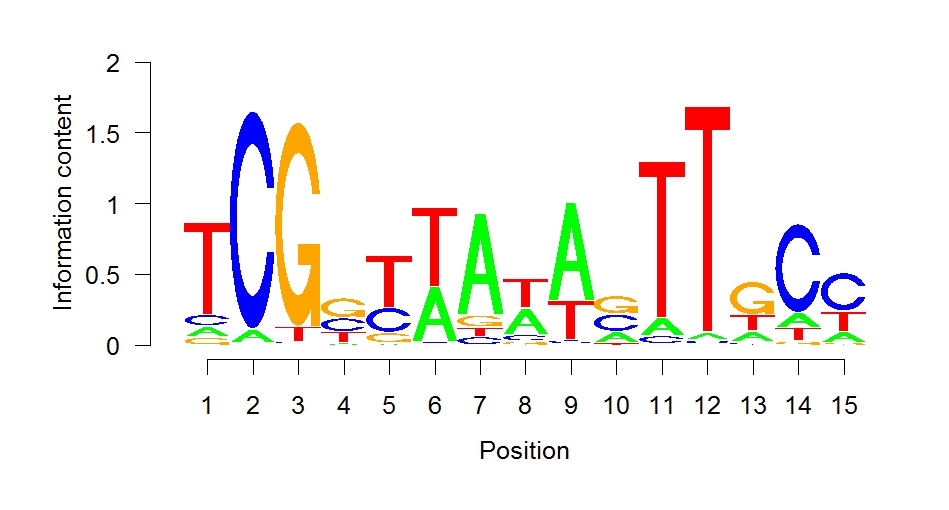
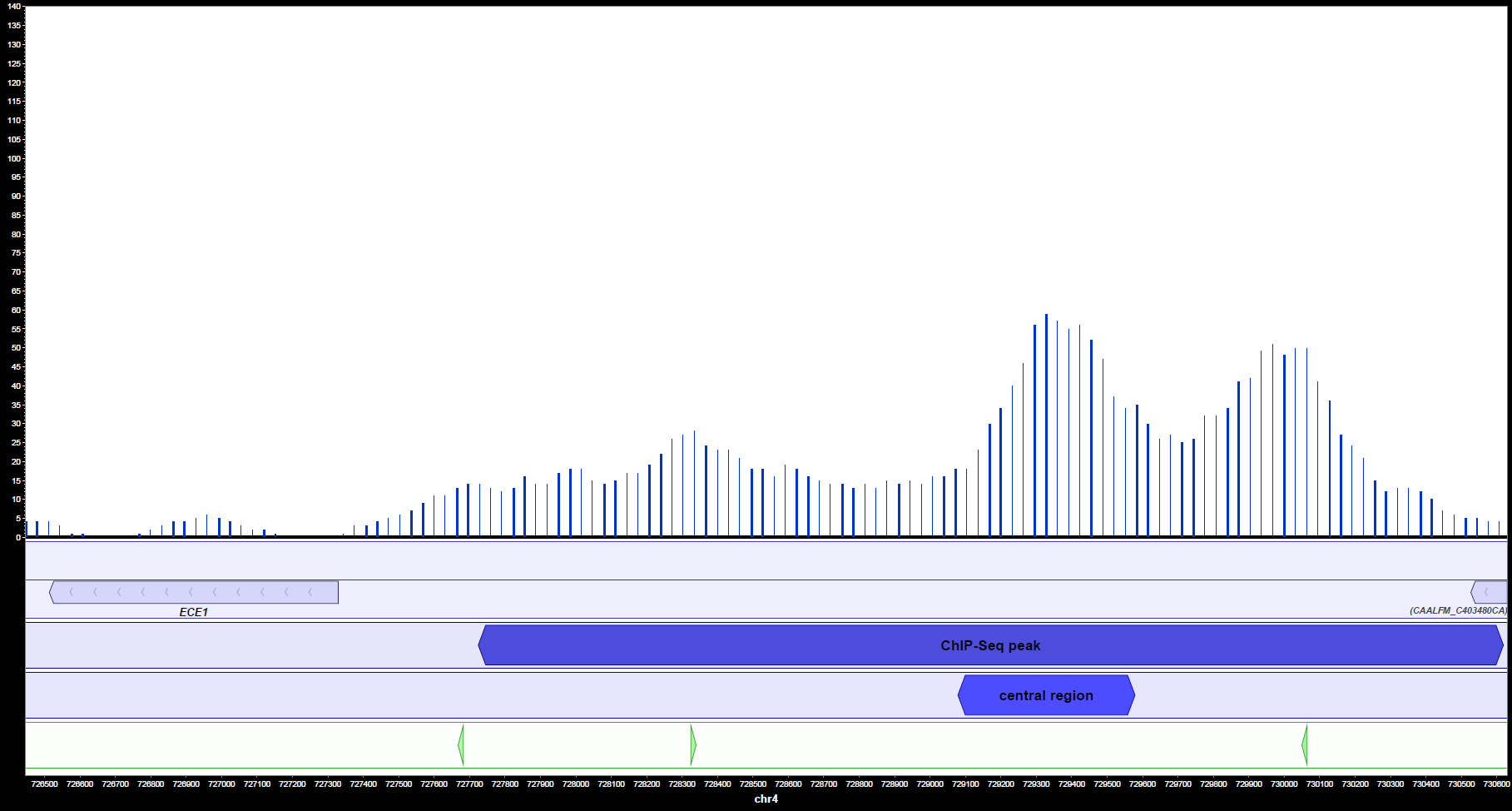
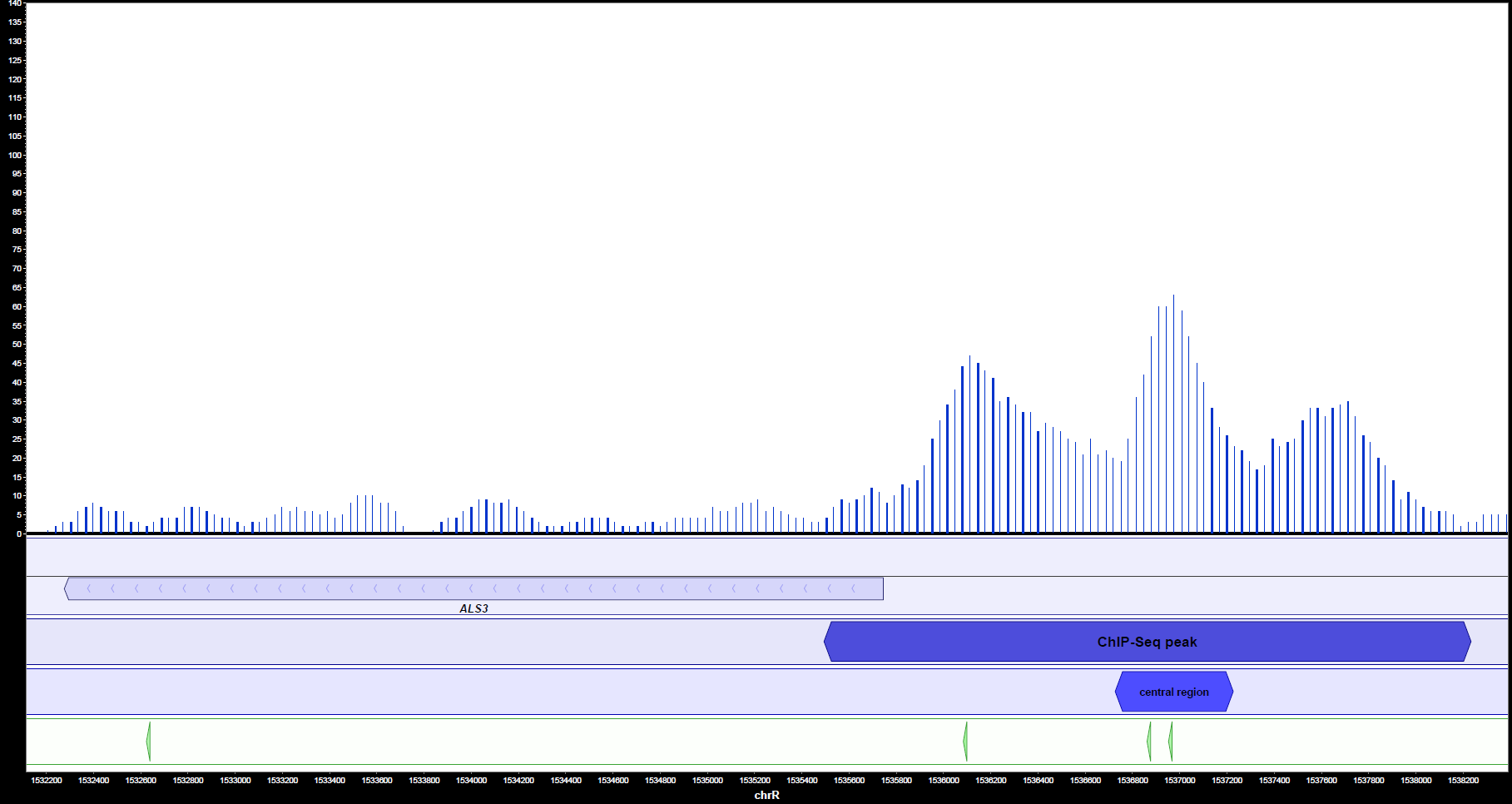
MochiView can also be used for visualization of peaks, genes and motifs. It shows clearly (figures 2a and 2b) that Ahr1 binds within the promoter region of important virulence associated genes of C. albicans like ECE1 and ALS3. Frequently, there is more than one potential binding site present in one promoter region.
Altogether, the binding motif was detected in the promoters of 37 virulence relevant genes. The table below shows the motif with the highest MochiView score for each gene. In case there are two motifs with the same score, both are shown.
| Gene Name | Motif Distance to Gene in bp | Motif Strand | Motif Score | Motif Sequence |
|---|---|---|---|---|
| AHR1 | 5586 | - | 5.4 | GGCAACAATTACCGG |
| ALS1 | 1390 | - | 4.6 | GGAAACTTCAAACGA |
| ALS3 | 340 | + | 4.5 | TGCAAGTTAAACCGA |
| ALS4 | 1649 | - | 3.4 | CACAAGTGTAAGCGA |
| BCR1 | 2178 | - | 2.8 | AGAAAGGAAAAGCGA |
| BRG1 | 5765 | + | 5.1 | TGCAAGAATTACCGA |
| CDR1 | 1337 | + | 4.7 | ATCAACTATTGCCGA |
| CZF1 | 4304 | - | 2.4 | TGCAGTGGTAACCGA |
| DCK1 | 506 | + | 4.5 | GGAAAGTATAGTCGA |
| DEF1 | 1719 | - | 4.0 | GTCAACTTCTGACGA |
| DEF1 | 495 | - | 4.0 | GGAAATTAGAAACGA |
| EAP1 | 1204 | + | 4.5 | GGGAAGTTCAAGCGA |
| ECE1 | 2721 | + | 4.8 | GGGAAGAATTACCGA |
| EFG1 | 2311 | + | 5.0 | TGCAACTACAACCGA |
| FLO8 | 2066 | - | 3.5 | GGCAAGAAGTAGAGA |
| HGC1 | 11647 | - | 4.3 | GGAAAGTGGTAGCGA |
| HGT2 | 3442 | + | 4.9 | TGCAACTATTCGCGA |
| HWP1 | 1225 | - | 3.9 | GGCAAGTTTATCCGC |
| HYR1 | 1938 | + | 4.8 | AGCAATATTAGGCGA |
| IHD1 | 861 | + | 5.1 | TGCAACAATTACCGA |
| LMO1 | 179 | + | 4.4 | AGAAATTTTTAGCGA |
| MDR1 | 537 | - | 5.4 | GGTAACTATTGGCGA |
| NDT80 | 1567 | - | 5.1 | GGCAAGTTTAATCGA |
| RBT1 | 98 | - | 4.4 | GGTAAGATTTACCGG |
| RIM101 | 664 | - | 5.1 | AGCAAGTAGAGCCGA |
| SAP4 | 807 | - | 3.8 | AGCAATTTTAAGAGA |
| SAP5 | 1144 | + | 4.3 | GGCAATTTTAAGAGA |
| SAP6 | 833 | - | 3.5 | GGTAATTTTAAGAGA |
| SFL1 | 6551 | - | 5.1 | GGAAACTATTACCGG |
| SFL2 | 2733 | + | 4.0 | AACAAGTAGAGCCGA |
| SOD5 | 888 | + | 4.9 | GGCATCTTTTCCCGA |
| SOD5 | 1512 | + | 4.9 | GGAAAGTTGAAGCGA |
| STP2 | 915 | + | 2.9 | TGCAAGACTTGCAGA |
| TEC1 | 5005 | - | 4.8 | GGCAAGTATAAGCTA |
| UME6 | 15928 | + | 4.2 | AACAACTTTAAACGA |
| WOR1 | 5648 | - | 5.0 | AGCAAGTATAGCCGT |
| WOR2 | 1834 | + | 5.0 | GGCATCAATTACCGA |
This indicates that Ahr1 is involved in the regulation of a multitude of virulence associated processes like hyphae formation, cell invasion, iron acquisition or host cell damage. Since the promoter of Ahr1 also shows the binding motif, it may regulate itself via a feedback loop.
Homology and Synteny
Homologues of AHR1 (genes which have the same precursor gene), but also of ECE1 and ALS3 can be found in the genomes of C. dubliniensis and C. tropicalis. Both species are close relatives of C. albicans and pathogens. The AHR1 homologues show a very good alignment score of 992 (C. albicans AHR1 vs. C. dubliniensis Cd36_85930) and 914 (C. albicans AHR1 vs. C. tropicalis CTRG_02263) in T-Coffee (v. 11.0) [7]. ECE1 und its homologues show scores of 994 (C. albicans ECE1 vs. C. dubliniensis Cd36_43260) and 852 (C. albicans ECE1 vs. C. tropicalis CTRG_00476).
Also, the spatial order of the genes in the genome (synteny) in all three organisms is similar, as can be visualized in the Candida Gene Order Browser [8], while it is quite different to that in S. cerevisiae, the non-pathogenic fungal model organism.
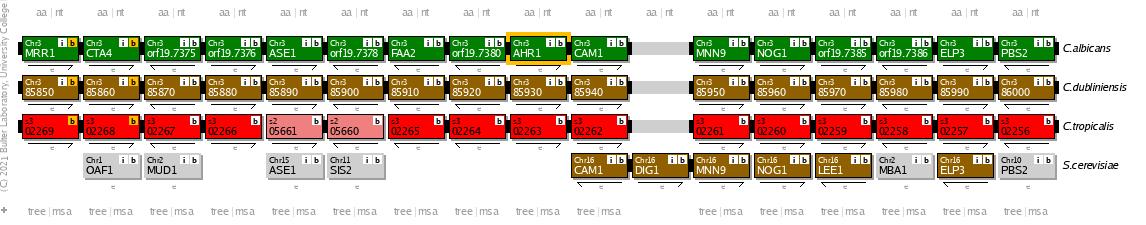
Therefore, the regulatory mechanisms described here could be conserved in those two species, too.
References
- [1] P. E. Sudbery: Growth of Candida albicans hyphae. In: Nat. Rev. Microbiol., 9:737–748, 2011. doi: 10.1038/nrmicro2636
- [2] K. Zakikhany, J. R. Naglik, A. Schmidt-Westhausen, G. Holland, M. Schaller, B. Hube: In vivo transcript profiling of Candida albicans identifies a gene essential for interepithelial dissemination. In: Cell Microbiol., 9:2938–2954, 2007. doi: 10.1111/j.1462-5822.2007.01009.x
- [3] A. L. Mavor, S. Thewes, B. Hube: Systemic fungal infections caused by Candida species: epidemiology, infection process and virulence attributes. In: Curr. Drug Targets, 6:863–874, 2005. doi: 10.2174/138945005774912735
- [4] S. Ruben, E. Garbe, S. Mogavero, D. Albrecht-Eckardt, D. Hellwig, A. Häder, T. Krüger, K. Gerth, I. D. Jacobsen, O. Elshafee, S. Brunke, K. Hünniger, O. Kniemeyer, A. A. Brakhage, J. Morschhäuser, B. Hube, S. Vylkova, O. Kurzai, R. Martin: Ahr1 and Tup1 contribute to the transcriptional control of virulence-associated genes in Candida albicans. In: mBio, 11:2, 2020.
doi: 10.1128/mBio.00206-20 - [5] P. Machanick, T. L. Bailey: MEME-ChIP: motif analysis of large DNAdatasets. In: Bioinformatics, 27:1696–1697, 2011. doi: 10.1093/bioinformatics/btr189
- [6] O. R. Homann, A. D. Johnson: MochiView: versatile software for genome browsing and DNA motif analysis. In: BMC Biol., 8:49, 2010. doi: 10.1186/1741-7007-8-49
- [7] C. Notredame, D. G. Higgins, J. Heringa: T-Coffee: A novel method for fast and accurate multiple sequence alignment. In: J. Mol. Biol., 302(1):205–217, 2000. doi: 10.1006/jmbi.2000.4042
- [8] S. L. Maguire, S. S. ÓhÉigeartaigh, K. P. Byrne, M. S. Schröder, P. O'Gaora, K. H. Wolfe, G. Butler: Comparative genome analysis and gene finding in Candida species using CGOB. In: Mol. Biol. Evol., 30(6):1281–1291, 2013. doi: 10.1093/molbev/mst042
